Track
GitHub Actions and MakeFile: A Hands-on Introduction
As data scientists, we usually don’t get involved with deployment and maintenance—we build the statistical models, and engineers do the rest.
However, things are starting to change!
With the increasing demand for data scientists who can bridge the gap between model creation and production, familiarizing yourself with automation and CI/CD (Continuous Integration/Continuous Deployment) tools can be a strategic advantage.
In this tutorial, we’ll learn about two popular automation tools: Make (local automation) and GitHub Actions (cloud-based automation). Our main focus will be on incorporating these tools into our data projects.

GitHub Action and Make. Image by Abid Ali Awan.
If you want to learn more about automation in the context of data science, check out this course on Fully Automated MLOps.
Introducing MakeFile
A Makefile is a blueprint for building and managing software projects. It is a file containing instructions for automating tasks, streamlining complex build processes, and ensuring consistency.
To execute the commands within the Makefile, we use the make command-line tool. This tool can be run like a Python program by providing the arguments or just running it on its own.
Core components
A Makefile usually consists of a list of targets, dependencies, actions, and variables:
- Target: These are the desired outcomes, like "build," "test," or "clean." Imagine them as goals you want to achieve through the Makefile.
- Dependencies: These are the files needed to build the target. Think of them as the ingredients required for each recipe (target).
- Actions: These are the instructions or scripts that tell the system how to build the target. They could involve commands like
python test.py. - Variables: Makefiles provide global variables that act like arguments passed to the actions. Using the action example above, we could have
python test.py $variable_1 $variable_2.
In a nutshell, this is how the template of a MakeFile could look like:
Variable = some_value
Target: Dependencies
Actions (commands to build the target) $VariableUsing our examples above, the MakeFile could look like this:
# Define variables
variable_1 = 5
variable_2 = 10
# Target to run a test with variables
test: test_data.txt # Dependency on test data file
python test.py $variable_1 $variable_2Installing the Make CLI Tool
We can install the Make CLI tool on all operating systems.
Linux
For Linux, we need to use the following command in the terminal:
$ sudo apt install makeMacOS
For macOS, we can use homebrew to install make:
$ brew install makeWindows
Windows is a bit different. The Make tool can be used in multiple ways. The most popular options are WSL (Ubuntu Linxus), w64devkit, and GnuWin32.
Let’s download and install GnuWin32 from the SourceForge page.
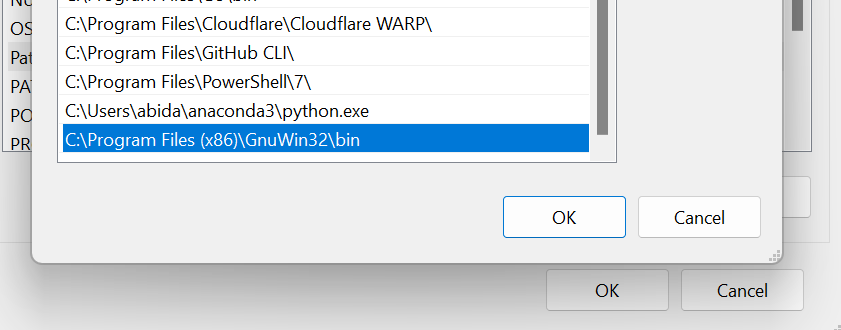
To use it as a command-line tool in the terminal, we need to add the folder's location to the Windows environment variable. These are the steps we can take:
- Search for "environment variables" in the start menu—this will redirect us to the System Properties window.
- Inside the System Properties window, click on the Environment Variables button.
- In the system variable section, locate the "Path" variable and edit it—this will launch another window where we can add the new path to the Make tool directory.
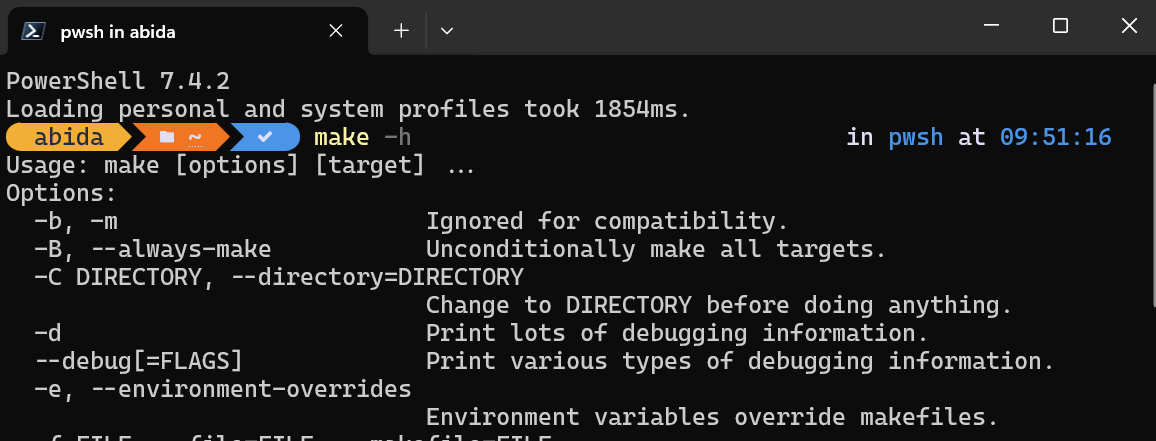
To test if Make is successfully installed, we can type make -h in the PowerShell.
Using MakeFile for a Data Science Project
In this section, we’ll learn how to use Makefile and Make CLI tools in a data science project.
We’ll use the World Happiness Report 2023 dataset to cover data processing, analysis, and saving data summaries and visualizations.
Setting up
We start by going to the project directory and creating a folder called Makefile-Action. We then create the data_processing.py file within that folder and launch the VSCode editor.
$ cd GitHub
$ mkdir Makefile-Action
$ cd .\Makefile-Action\
$ code data_processing.pyData processing
We begin with processing our data using Python. In the code block below, we take the following steps (note: this is an example code—in a real-world scenario, we’d probably have to perform more steps):
- Load the raw dataset from the directory location provided by the user.
- Rename all the columns for better readability.
- Fill in the missing values.
- Save the cleaned dataset.
File: data_processing.py
import sys
import pandas as pd
# Check if the data location argument is provided
if len(sys.argv) != 2:
print("Usage: python data_processing.py <data_location>")
sys.exit(1)
# Load the raw dataset (step 1)
df = pd.read_csv(sys.argv[1])
# Rename columns to more descriptive names (step 2)
df.columns = [
"Country",
"Happiness Score",
"Happiness Score Error",
"Upper Whisker",
"Lower Whisker",
"GDP per Capita",
"Social Support",
"Healthy Life Expectancy",
"Freedom to Make Life Choices",
"Generosity",
"Perceptions of Corruption",
"Dystopia Happiness Score",
"GDP per Capita",
"Social Support",
"Healthy Life Expectancy",
"Freedom to Make Life Choices",
"Generosity",
"Perceptions of Corruption",
"Dystopia Residual",
]
# Handle missing values by replacing them with the mean (step 3)
df.fillna(df.mean(numeric_only=True), inplace=True)
# Check for missing values after cleaning
print("Missing values after cleaning:")
print(df.isnull().sum())
print(df.head())
# Save the cleaned and normalized dataset to a new CSV file (step 4)
df.to_csv("processed_data\WHR2023_cleaned.csv", index=False)Data analysis
Now we continue with analyzing the data and save all our code into a file named data_analysis.py. We can create this file with the following terminal command:
$ code data_analysis.pyWe can also use the echo command to create the Python file and open it in a different code editor.
$ echo "" > data_analysis.pyIn the code block below, we:
- Load the clean dataset using the input provided by the user.
- Generate the data summary, the data information, and the first 5 rows of the DataFrame and save it in the TXT file.
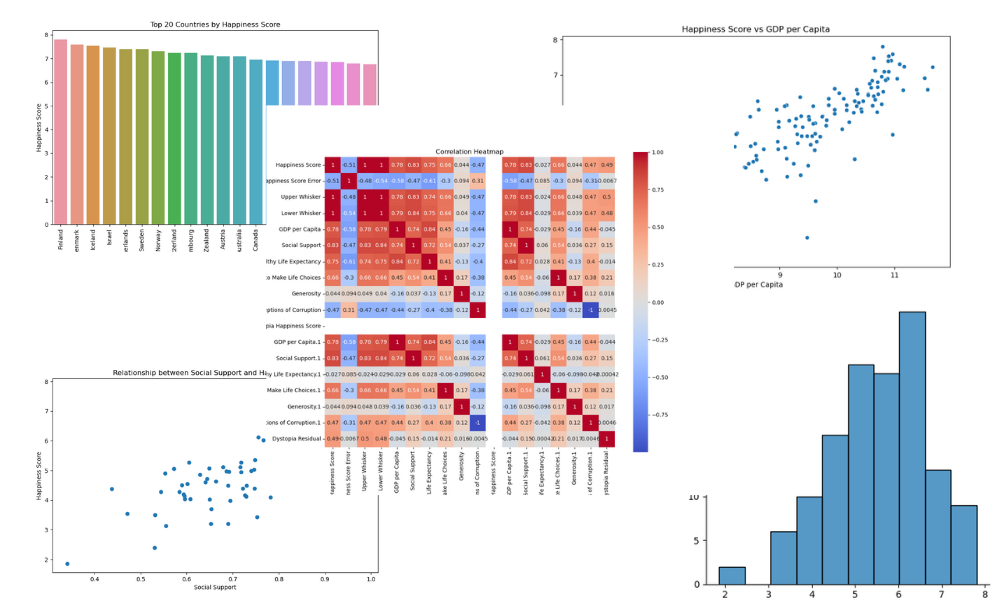
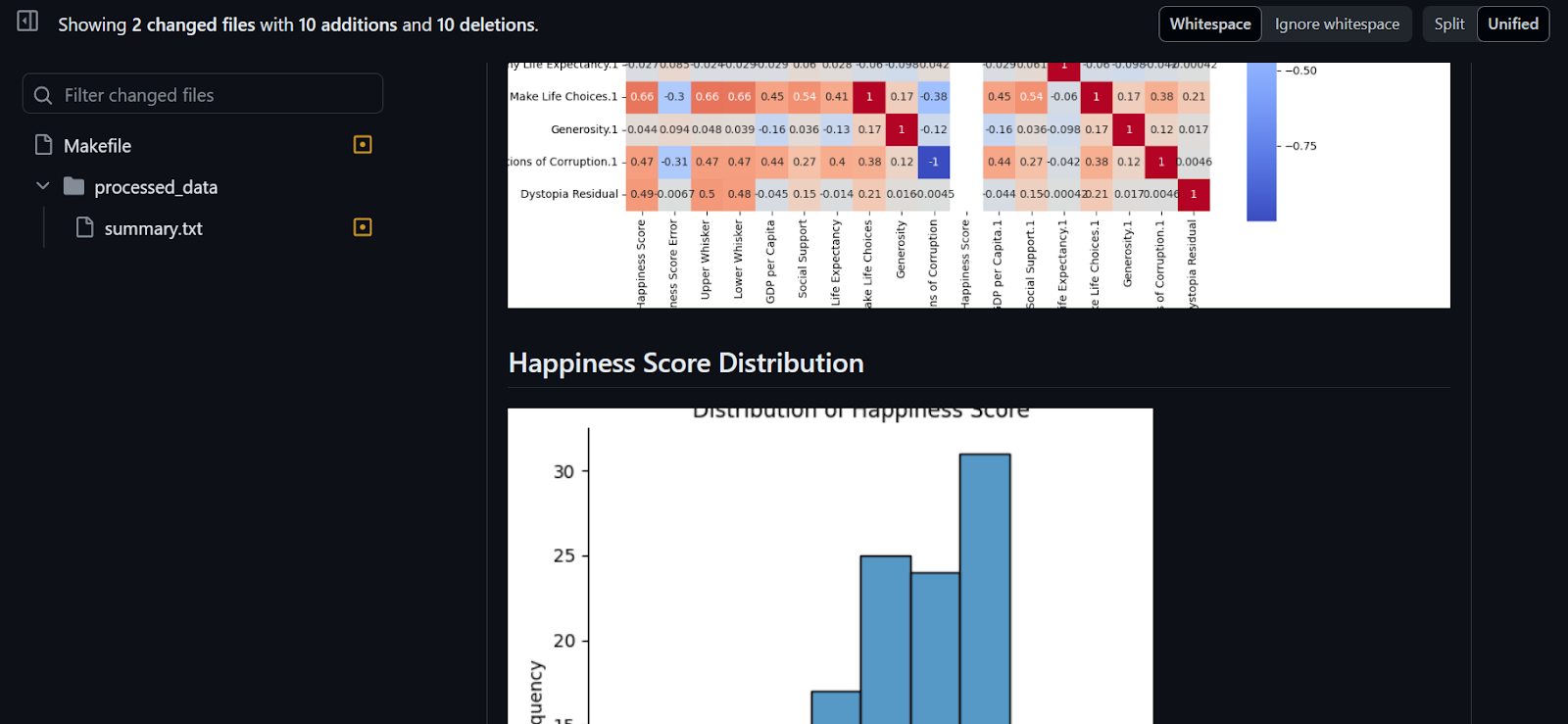
- Plot visualization for:
- The distribution of happiness scores
- Top 20 countries by happiness score
- Happiness score versus GDP per capita
- Happiness Score versus social support
- A correlation heatmap
- Save all the visualizations in the “figures” folder.
File: data_analysis.py
import io
import sys
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
# Check if the data location argument is provided
if len(sys.argv) != 2:
print("Usage: python data_analysis.py <data_location>")
sys.exit(1)
# Load the clean dataset (step 1)
df = pd.read_csv(sys.argv[1])
# Data summary (step 2)
print("Data Summary:")
summary = df.describe()
data_head = df.head()
print(summary)
print(data_head)
# Collecting data information
buffer = io.StringIO()
df.info(buf=buffer)
info = buffer.getvalue()
## Write metrics to file
with open("processed_data/summary.txt", "w") as outfile:
f"\n## Data Summary\n\n{summary}\n\n## Data Info\n\n{info}\n\n## Dataframe\n\n{data_head}"
print("Data summary saved in processed_data folder!")
# Distribution of Happiness Score (step 3)
plt.figure(figsize=(10, 6))
sns.displot(df["Happiness Score"])
plt.title("Distribution of Happiness Score")
plt.xlabel("Happiness Score")
plt.ylabel("Frequency")
plt.savefig("figures/happiness_score_distribution.png")
# Top 20 countries by Happiness Score
top_20_countries = df.nlargest(20, "Happiness Score")
plt.figure(figsize=(10, 6))
sns.barplot(x="Country", y="Happiness Score", data=top_20_countries)
plt.title("Top 20 Countries by Happiness Score")
plt.xlabel("Country")
plt.ylabel("Happiness Score")
plt.xticks(rotation=90)
plt.savefig("figures/top_20_countries_by_happiness_score.png")
# Scatter plot of Happiness Score vs GDP per Capita
plt.figure(figsize=(10, 6))
sns.scatterplot(x="GDP per Capita", y="Happiness Score", data=df)
plt.title("Happiness Score vs GDP per Capita")
plt.xlabel("GDP per Capita")
plt.ylabel("Happiness Score")
plt.savefig("figures/happiness_score_vs_gdp_per_capita.png")
# Visualize the relationship between Happiness Score and Social Support
plt.figure(figsize=(10, 6))
plt.scatter(x="Social Support", y="Happiness Score", data=df)
plt.xlabel("Social Support")
plt.ylabel("Happiness Score")
plt.title("Relationship between Social Support and Happiness Score")
plt.savefig("figures/social_support_happiness_relationship.png")
# Heatmap of correlations between variables
corr_matrix = df.drop("Country", axis=1).corr()
plt.figure(figsize=(12, 10))
sns.heatmap(corr_matrix, annot=True, cmap="coolwarm", square=True)
plt.title("Correlation Heatmap")
plt.savefig("figures/correlation_heatmap.png")
print("Visualizations saved to figures folder!")Creating Makefile
Before we create a Makefile, we need to set up a requirements.txt file to install all required Python packages on a new machine. This is how the requirements.txt file will look like:
pandas
numpy
seaborn
matplotlib
blackOur Makefile consists of variables, dependencies, targets, and actions—we’ll learn all the building blocks. We create a file named Makefile and start adding the actions for the targets:
- install: This upgrades the pip version and installs all the necessary Python packages using the requirements.txt file.
- format: This formats all Python files using the Black tool.
- process: This runs the `data_processing.py` Python file with a raw data path as a variable and dependency.
- analyze: This runs the `data_analysis.py` Python file with a processed data path as a variable and dependency.
- clean: This removes all the files and figures generated while running the Python files.
- all: This runs all the valid targets in a sequence.
File: Makefile
RAW_DATA_PATH = "raw_data/WHR2023.csv"
PROCESSED_DATA = "processed_data/WHR2023_cleaned.csv"
install:
pip install --upgrade pip &&\
pip install -r requirements.txt
format:
black *.py --line-length 88
process: ./raw_data/WHR2023.csv
python data_processing.py $(RAW_DATA_PATH)
analyze: ./processed_data/WHR2023_cleaned.csv
python data_analysis.py $(PROCESSED_DATA)
clean:
rm -f processed_data/* **/*.png
all: install format process analyzeTo execute the target, we’ll use the make tool and provide the target name in the terminal.
$ make formatThe Black script ran successfully.
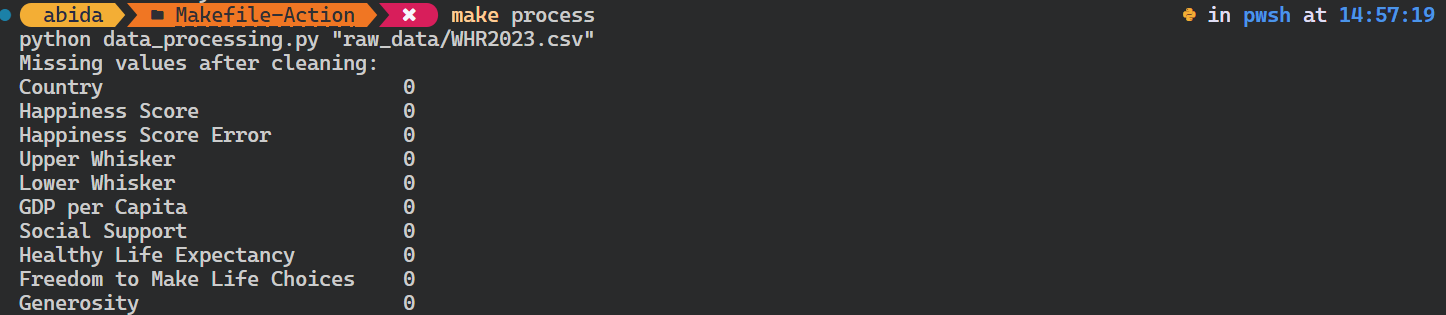
Let’s try to run the Python script for data processing.
It’s important to note that it checks the target dependencies before running the process target. In our case, it checks whether the raw data file exists. If it doesn’t, the command won’t initiate.
$ make processAs we can see, it is that simple.
You can even override the existing variable by providing an additional argument to the make command.
In our case, we have changed the path of the raw data.
$ make process RAW_DATA_PATH="WHR2022.csv"The Python script ran with the different input argument.
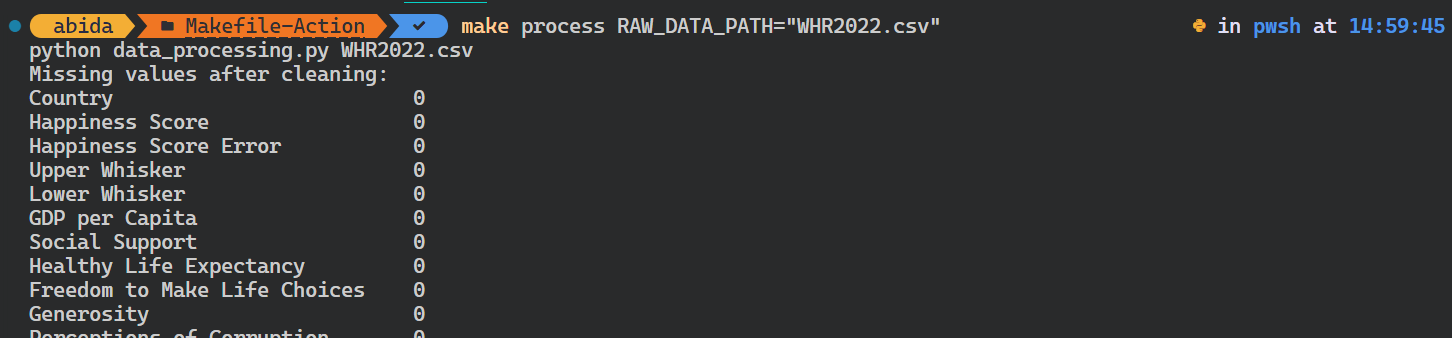
To automate the entire workflow, we’ll use all as a target, which will learn, install, format, process, and analyze targets one by one.
$ make allThe make command installed the Python packages, formatted the code, processed the data, and saved the summary and visualizations.
This is how your project directory should look like after running the make all command:
Introducing GitHub Actions
While Make excels at local automation within our development environment, GitHub Actions offers a cloud-based alternative.
GitHub Actions is generally used for CI/CD, which allows developers to compile, build, test, and deploy the application to production directly from GitHub.
For example, we can create a custom workflow that will be triggered based on specific events like push or pull requests. That workflow will run shell scripts and Python scripts, or we can even use pre-built actions. The custom workflow is a YML file, and it’s usually pretty straightforward to understand and start writing custom runs and actions.
Core components
Let’s explore the core components of GitHub Actions:
- Workflows are defined using YML files, which specify the steps that should be executed and the conditions under which they should be executed.
- Events are specific activities that trigger a workflow run like push, pull_request, schedule, and more.
- Runners are machines that run the workflows when they're triggered. GitHub provides hosted, free runners for Linux, Windows, and macOS.
- Jobs are a set of steps executed on the same runner. By default, jobs run in parallel but can be configured to run sequentially.
- Steps are tasks that run commands or actions similar to Makefile actions.
- Actions are custom applications for the GitHub Actions platform that perform a complex but frequently repeated task. There are many actions to select from, most of which are supported by the open-source community.
Getting Started with GitHub Actions
In this section, we’ll learn to use GitHub Actions and try to replicate the commands we previously used in the Makefile.
To transfer our code to GitHub, we have to convert our project folder to a Git repository using:
$ git initThen, we create a new repository in GitHub and copy the URL.
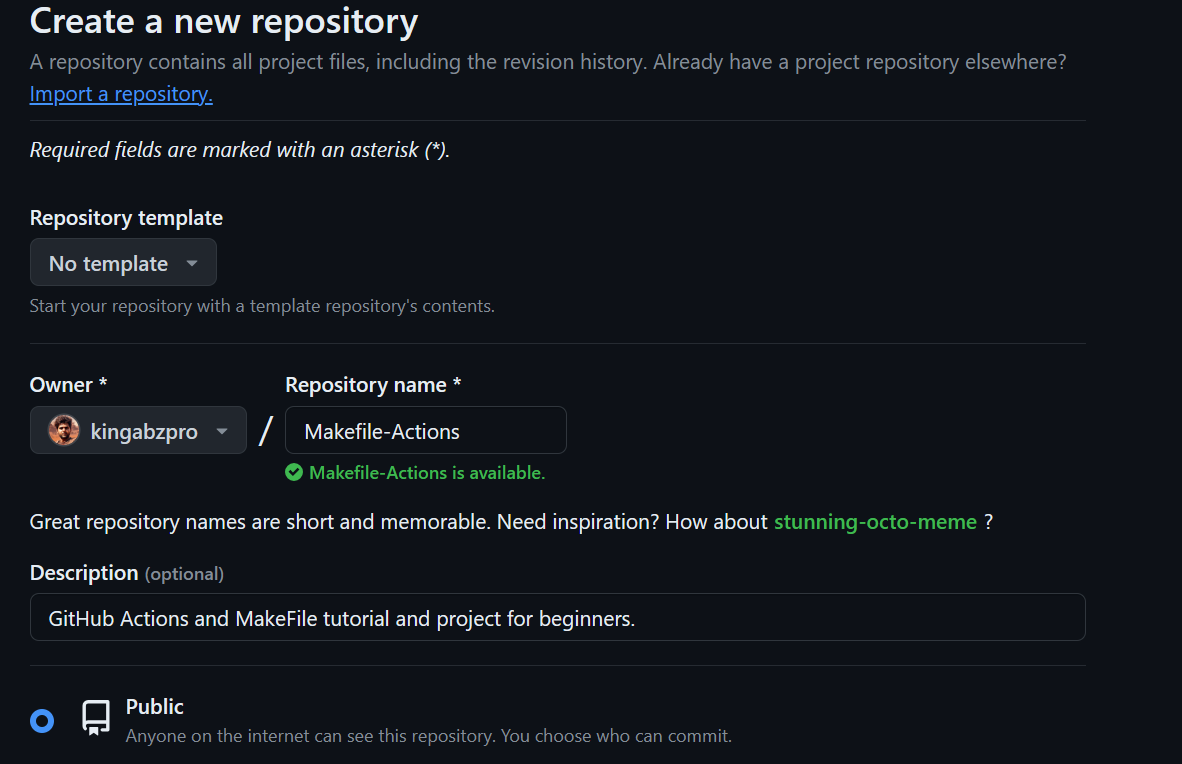
In the terminal, we type the commands below to:
- Add the remote repository link.
- Pull the files from the remote repository.
- Add and commit all the files with a commit message.
- Push the file from the local
masterto the remotemainbranch.
$ git remote add github https://github.com/kingabzpro/Makefile-Actions.git
$ git pull github main
$ git add .
$ git commit -m "adding all the files"
$ git push github master:mainAs a side note, if you need a brief recap on GitHub or Git, check out this beginner-friendly tutorial on GitHub and Git. It’ll help you version your data science project and share it with the team using the Git CLI tool.
Continuing our lesson—we see that all the files have been successfully transferred to the remote repository.
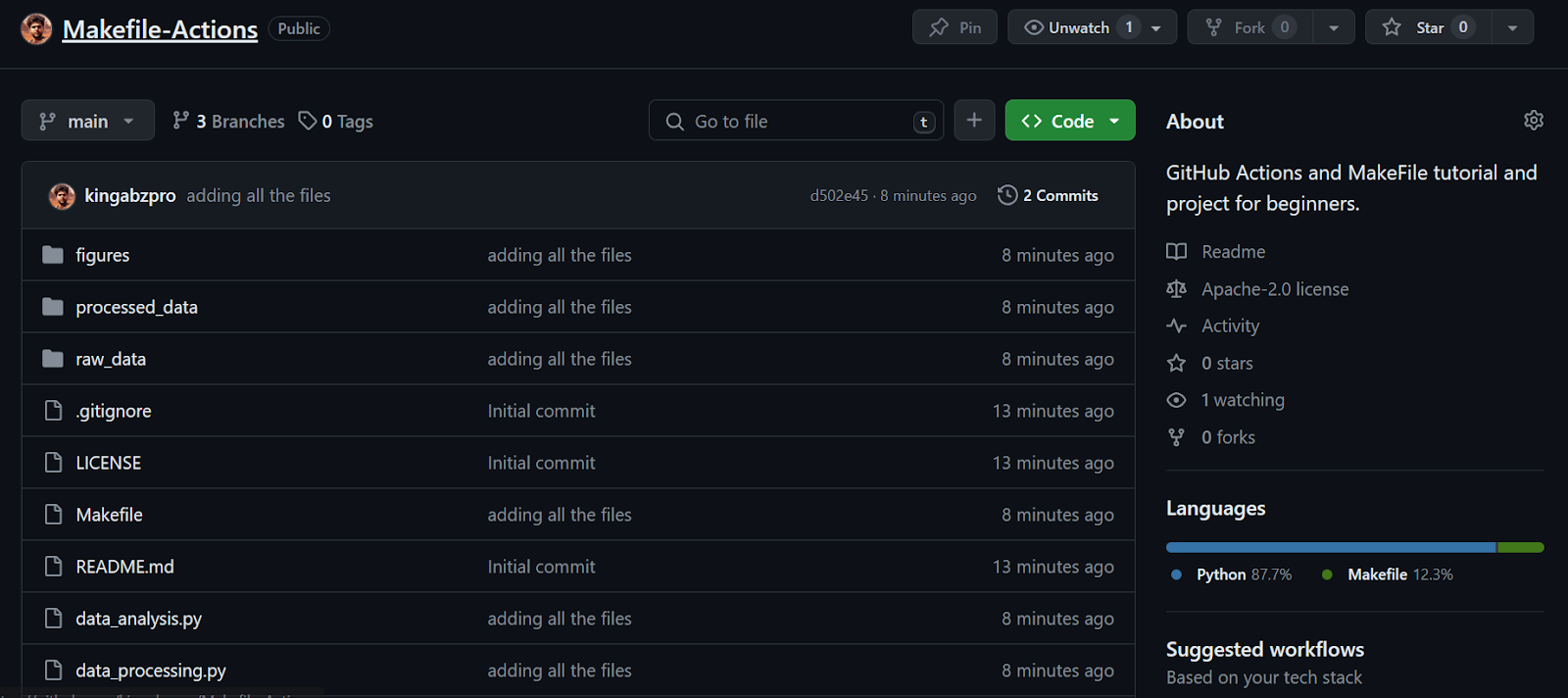
Next, we’ll create a GitHub Action workflow. We first need to go to the Actions tab on our repository kingabzpro/Makefile-Actions. Then, we click the blue text “set up a workflow yourself.”
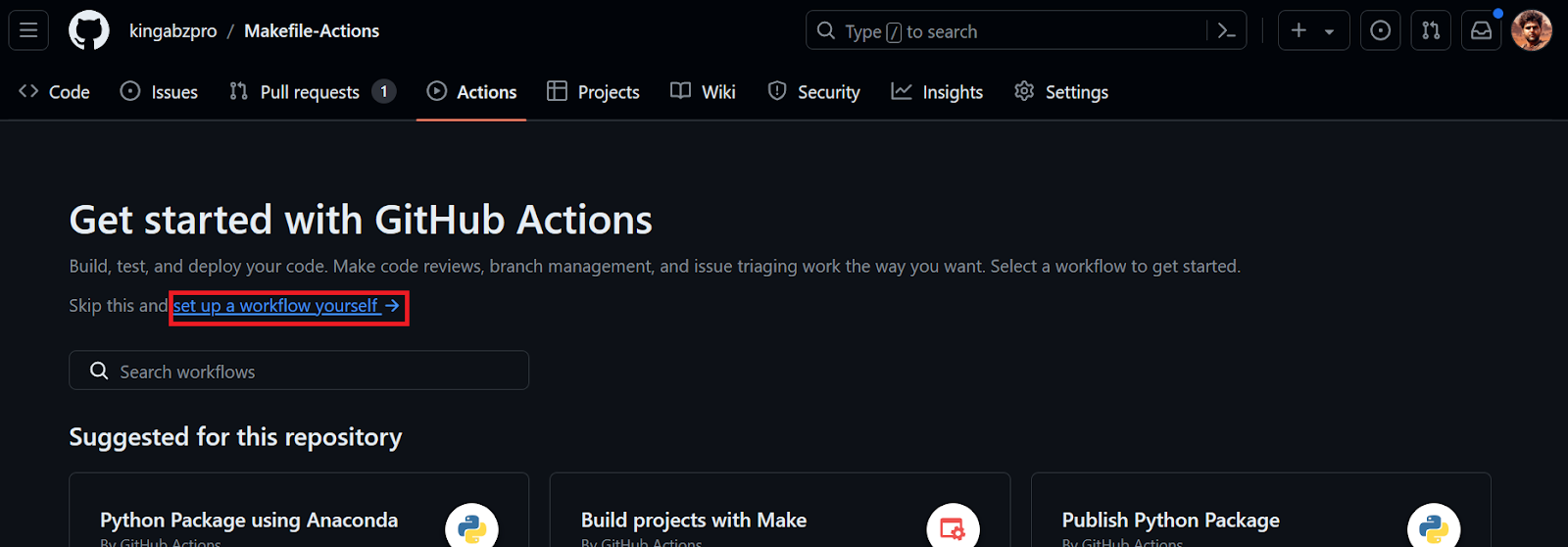
We’ll be redirected to the YML file, where we’ll write all the code for setting up the environment and executing the commands. We will:
- Provide the workflow name.
- Set up the trigger events to execute the workflow when code is pushed to the
mainbranch or there is a pull request on themainbranch. - Set up a job with the latest Ubuntu Linux machine.
- Use the checkout v3 action in the step section. It helps our workflow to access repositories.
- Install the necessary Python packages, format the code, and run both Python scripts with variables.
name: Data Processing and Analysis
on:
push:
branches: [ "main" ]
pull_request:
branches: [ "main" ]
jobs:
build:
runs-on: ubuntu-latest
steps:
- uses: actions/checkout@v3
- name: Install Packages
run: |
pip install --upgrade pip
pip install -r requirements.txt
- name: Format
run: black *.py --line-length 88
- name: Data Processing
env:
RAW_DATA_DIR: "./raw_data/WHR2023.csv"
run: python data_processing.py $RAW_DATA_DIR
- name: Data Analysis
env:
CLEAN_DATA_DIR: "./processed_data/WHR2023_cleaned.csv"
run: python data_analysis.py $CLEAN_DATA_DIRAfter committing the workflow file with the message, it’ll automatically start running the workflow.
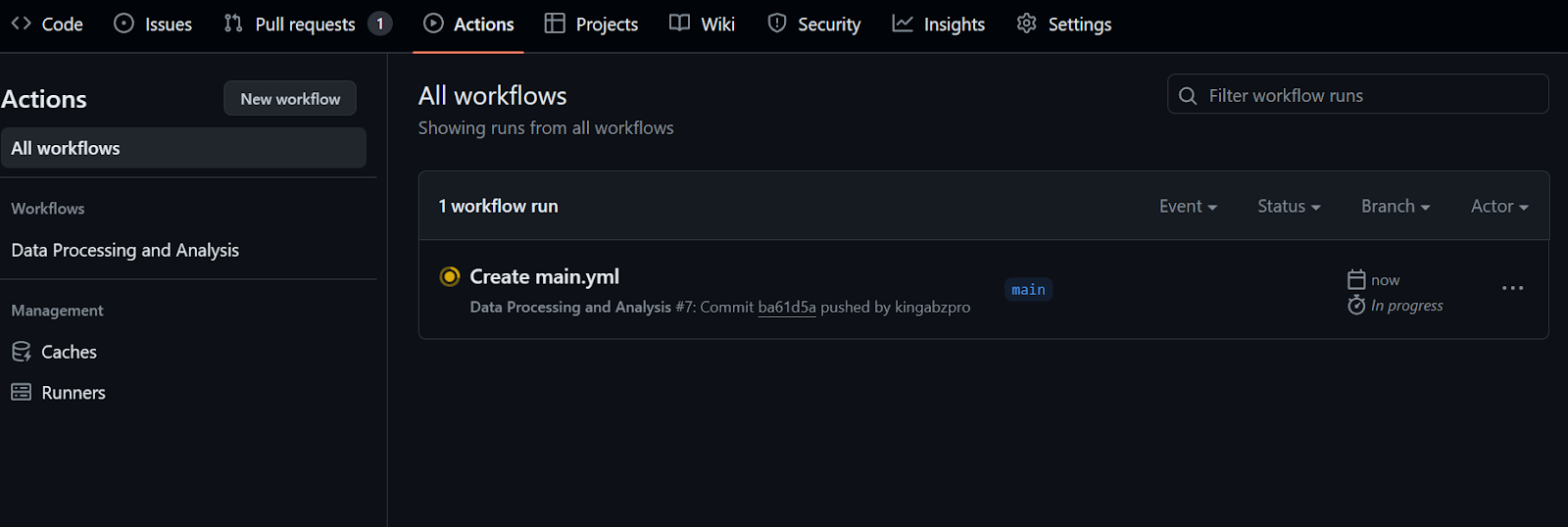
It will take at least 20 seconds to finish all the steps.
To check the logs, we first click on the workflow run, then the Build button, and then click on each job to access the logs.
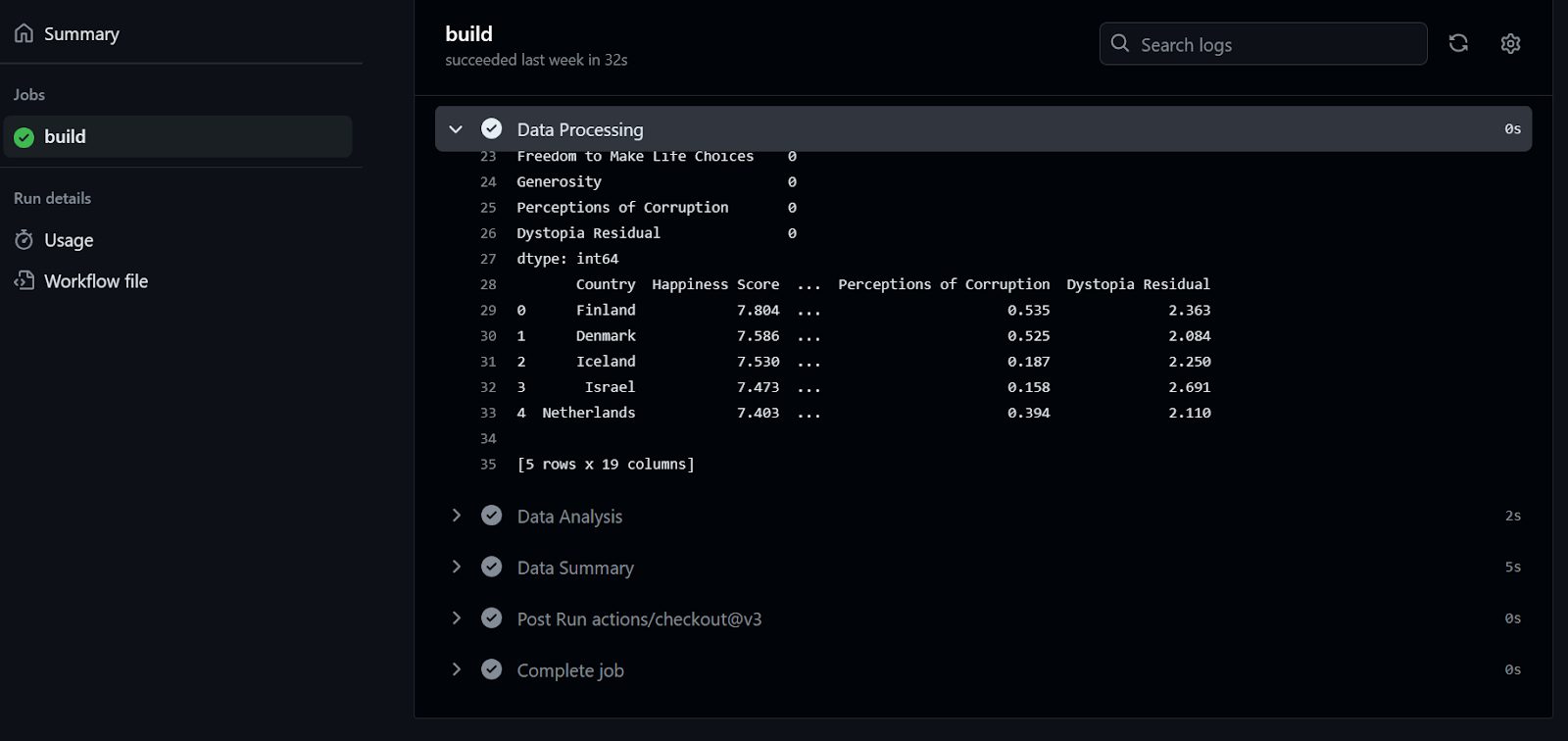
If this was fun for you, you will surely enjoy this Beginner's Guide to CI/CD for Machine Learning, which covers the basics of building and automating a machine learning workflow.
Combining GitHub Actions with MakeFile
To simplify and standardize the GitHub Action workflows, developers use Make commands within the workflow file. In this section, we’ll learn to simplify our workflow code using the Make command and learn to use the continuous machine learning (CML) action.
Before we start, we have to pull the workflow file from the remote repository.
$ git pullUsing CML
Continuous Machine Learning (CML) is an open-source library by iterative.ai that allows us to implement continuous integration within our data science project.
In this project, we’ll use the iterative/setup-cml GitHub Action that uses CML functions in the workflow to generate the analysis report using figures and data statistics.
The report will be created and attached to our GitHub commit for our team to review and approve before merging.
We’ll modify the Makefile and add another target “summary.” Note that:
- The target is a multi-line script that transfers the text data from summary.txt to the report.md.
- Then, one by one, it adds the headings for each figure and Markdown code to display the figure in the report.md file.
- In the end, we’ll use the
cmltool to create a data analytical report and display it under the commit comments.
Imagine adding so many lines to the GitHub workflow file—it’d be hard to read and modify. Instead, we’ll use make summary.
File: Makefile
summary: ./processed_data/summary.txt
echo "# Data Summary" > report.md
cat ./processed_data/summary.txt >> report.md
echo '\n# Data Analysis' >> report.md
echo '\n## Correlation Heatmap' >> report.md
echo '' >> report.md
echo '\n## Happiness Score Distribution' >> report.md
echo '' >> report.md
echo '\n## Happiness Score vs GDP per Capita' >> report.md
echo '' >> report.md
echo '\n## Social Support vs Happiness Relationship' >> report.md
echo '' >> report.md
echo '\n## Top 20 Countries by Happiness Score' >> report.md
echo '' >> report.md
cml comment create report.md
Modifying the GiHub Action file
We will now edit our main.yml file, which we can find in the .github/workflows directory.
We change all the commands and Python scripts with the make command and provide permission for the CML Action to create the data report in the commit comment. Additionally, let’s make sure we don’t forget to add the iterative/setup-cml@v3 Action to our run.
name: Data Processing and Analysis
on:
push:
branches: [ "main" ]
pull_request:
branches: [ "main" ]
permissions: write-all
jobs:
build:
runs-on: ubuntu-latest
steps:
- uses: actions/checkout@v3
- uses: iterative/setup-cml@v3
- name: Install Packages
run: make install
- name: Format
run: make format
- name: Data Processing
env:
RAW_DATA_DIR: "./raw_data/WHR2023.csv"
run: make process RAW_DATA_PATH=$RAW_DATA_DIR
- name: Data Analysis
env:
CLEAN_DATA_DIR: "./processed_data/WHR2023_cleaned.csv"
run: make analyze PROCESSED_DATA=$CLEAN_DATA_DIR
- name: Data Summary
env:
REPO_TOKEN: ${{ secrets.GITHUB_TOKEN }}
run: make summaryRunning the workflow
To execute the workflow, we only need to commit all the changes and sync them with the remote main branch.
$ git commit -am "makefile and github action combo"
$ git push github master:main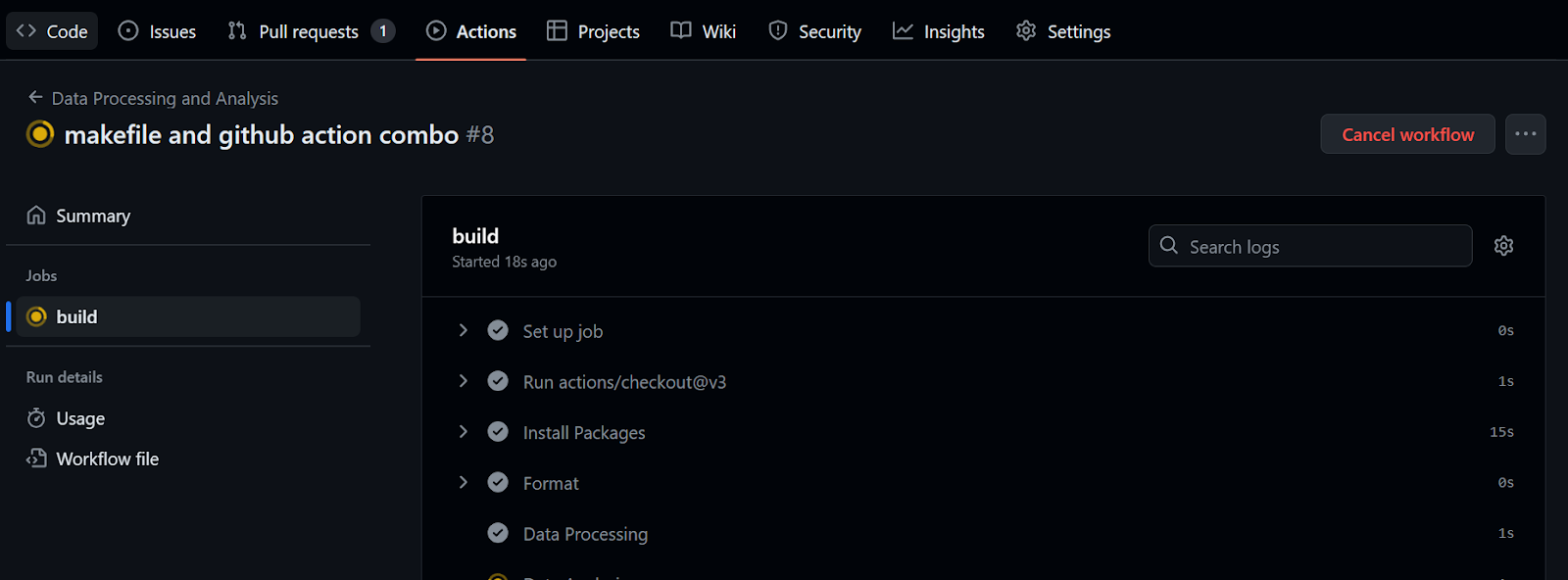
It took 32 seconds to run the workflow—our report is ready to be reviewed. To do that, we go to the build summary, scroll down to click on the Data Summary tab, and then click on the comment link (the link you see on line 20 in the figure below):
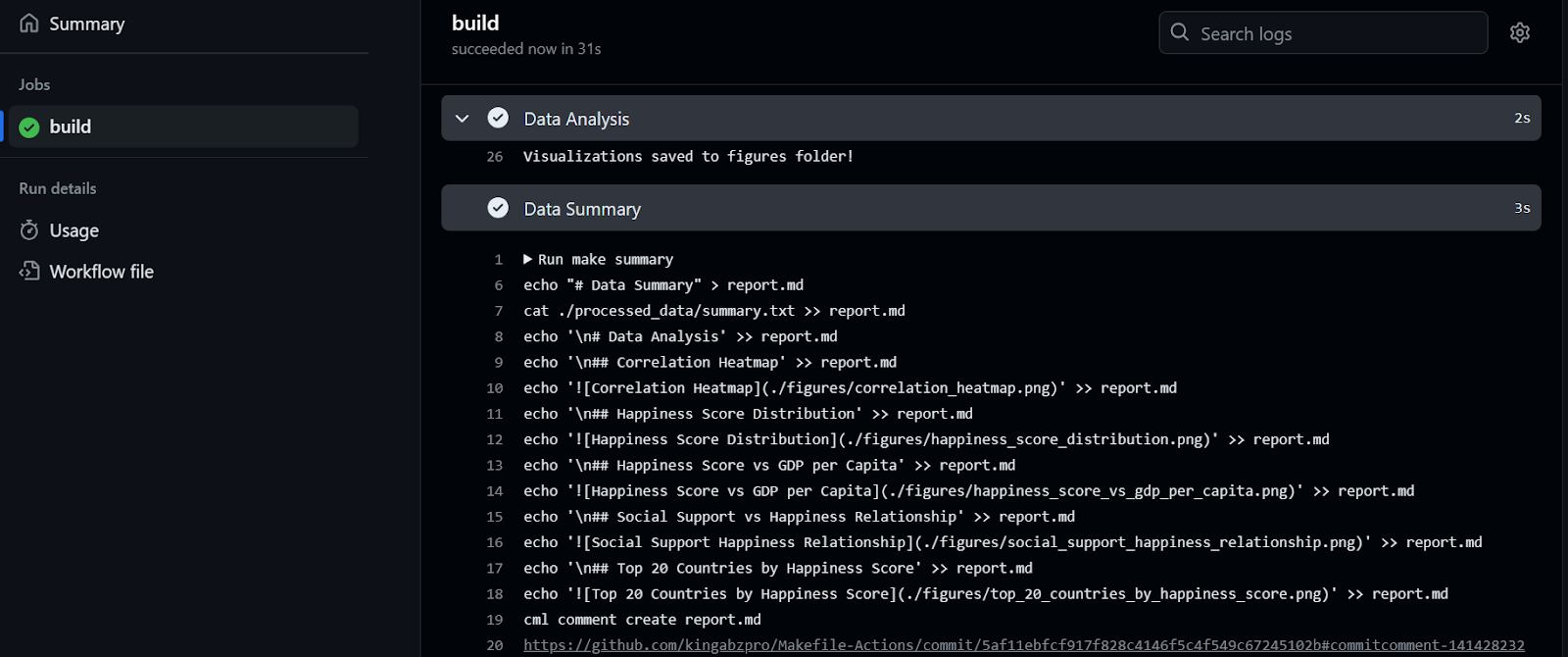
As we can see, the data summary and data visualizations are attached to our commit.
The project is available on kingabzpro/Makefile-Actions, and you can use it as a guide when you get stuck. The repository is public, so just fork it and experience the magic yourself!
Conclusion
In this tutorial, we focused on Makefile and GitHub Actions for automating the generation of data analytical reports.
We also learned how to build and run the workflow and use the Make tool within the GitHub workflow to optimize and simplify the automation process. Instead of writing multiple lines of code, we can use make summary to generate a data analysis report that will be attached to our commit.
If you want to master the art of CI/CD for data science, try out this course on CI/CD for Machine Learning. It covers the fundamentals of CI/CD, GitHub Actions, data versioning, and automating model hyperparameter optimization and evaluation.
I am a certified data scientist who enjoys building machine learning applications and writing blogs on data science. I am currently focusing on content creation, editing, and working with large language models.
Become a machine learning engineer!
Track
Data Engineer
Course
CI/CD for Machine Learning
blog
An Introduction to Data Orchestration: Process and Benefits
Srujana Maddula
9 min
blog
The Top 21 Airflow Interview Questions and How to Answer Them
Jake Roach
13 min
podcast
The Database is the Operating System with Mike Stonebraker, CTO & Co-Founder At DBOS
tutorial
Mastering AWS Step Functions: A Comprehensive Guide for Beginners
tutorial
Airflow vs Prefect: Deciding Which is Right For Your Data Workflow
Tim Lu
8 min
tutorial
Building an ETL Pipeline with Airflow
Jake Roach
15 min